消极的时候控制核孔侑dF4y2Ba
负控制RNAi实验孔侑dF4y2Ba
负控制RNAi实验孔侑dF4y2Ba

猫。不。/ ID:孔侑dF4y2Ba1027292孔侑dF4y2Ba

猫。不。/ ID:孔侑dF4y2Ba1027295孔侑dF4y2Ba

猫。不。/ ID:孔侑dF4y2BaSI03650318孔侑dF4y2Ba

猫。不。/ ID:孔侑dF4y2Ba1027281孔侑dF4y2Ba

猫。不。/ ID:孔侑dF4y2Ba1027280孔侑dF4y2Ba
消极的时候控制核的性能通过实验验证表所示。孔侑dF4y2Ba
| 测试类型孔侑dF4y2Ba | 测试的名字孔侑dF4y2Ba | 目的孔侑dF4y2Ba | 结果为负的时候控制核孔侑dF4y2Ba |
|---|---|---|---|
| 全基因组分析孔侑dF4y2Ba | Affymetrix GeneChip数组孔侑dF4y2Ba | 非特异性调节基因的表达孔侑dF4y2Ba | 最小数量的基因调控孔侑dF4y2Ba |
| 细胞试验孔侑dF4y2Ba | Live-cell细胞核染色孔侑dF4y2Ba | 核大小孔侑dF4y2Ba | 正常的孔侑dF4y2Ba |
| 细胞试验孔侑dF4y2Ba | 手机号孔侑dF4y2Ba | 扩散率孔侑dF4y2Ba | 不变孔侑dF4y2Ba |
| 细胞试验孔侑dF4y2Ba | 核苷酸结合孔侑dF4y2Ba | DNA合成速率孔侑dF4y2Ba | 不变孔侑dF4y2Ba |
| 细胞试验孔侑dF4y2Ba | Live-cell染料排斥孔侑dF4y2Ba | 细胞毒性作用孔侑dF4y2Ba | 不变孔侑dF4y2Ba |
| 细胞试验孔侑dF4y2Ba | 脱氧核糖核酸染色孔侑dF4y2Ba | 细胞循环分布孔侑dF4y2Ba | 正常的孔侑dF4y2Ba |
| RISC-incorporation分析孔侑dF4y2Ba(海拉和MCF-7细胞)孔侑dF4y2Ba | 记者构造转染孔侑dF4y2Ba | 确定是否纳入RISC核(有效负控制应该输入RISC)孔侑dF4y2Ba | 纳入RISC孔侑dF4y2Ba |
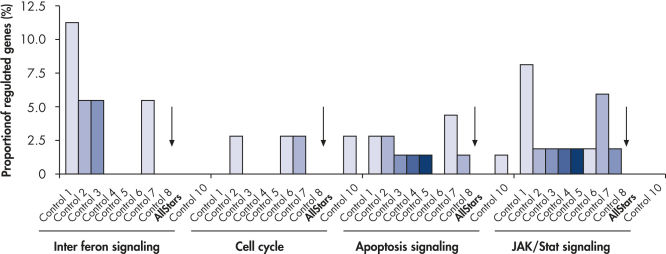
全基因组分析测试水平的非特异性影响转染多种负控制siRNAs后基因表达。从不同的起源被转染到多个负控制siRNAs MCF-7, K562,主要使用HiPerFect HUVEC细胞转染试剂。其中包括nonsilencing siRNAs(没有哺乳动物基因的同源性),炒siRNAs (siRNAs相同的碱基组成gene-specific siRNA但改变序列),和siRNAs目标人工报告基因。随后,整个人类基因组的表达谱进行了Affymetrix GeneChip数组。时候负控制核一致导致是非监管的最低数量的基因,使其高度合适的负控制。相比之下,其他负面控制siRNAs导致非特异性调节许多基因的重要的细胞通路(参见图”孔侑dF4y2Ba Multiple negative control siRNAs (Control 1– Control 10) were transfected in triplicate into MCF-7 cells. After incubation, cRNA was prepared and hybridized to Affymetrix human U133 GeneChip arrays. Regulated genes were identified as genes that showed at least a 1.5-fold change in expression (both upregulated and downregulated) compared to untransfected cells. Ingenuity pathway analysis software was used to determine the proportion of regulated genes in each pathway compared to the total number of genes identified as central to that pathway. Where a bar appears in the figure, this means that genes in the pathway were regulated by the siRNA. If every pathway gene was regulated, the relative proportion would be 100%. Lower bars therefore indicate a lower relative proportion of regulated genes within that pathway. Where no bar appears, no genes of the pathway were regulated by the siRNA. AllStars Negative Control siRNA (indicated with arrow) resulted in the lowest number of regulated genes. In contrast, other control siRNAs resulted in higher numbers of regulated genes from important cellular pathways.Low nonspecific effects on expression.
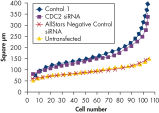
Live-cell细胞核染色是用来测量核大小。大小的变化可能是一个迹象表明细胞循环干扰或抑制增长。一系列的负控制siRNAs不同类型进行了测试。时候负控制核提供了最好的结果的所有控制测试。转染的时候负控制核没有导致任何改变核大小与untransfected细胞。相比之下,转染的另一个消极控制核(控制1)导致扩大核(参见图”孔侑dF4y2Ba HCT 116 cells were transfected with AllStars Negative Control siRNA, another negative control siRNA (Control 1), and CDC2 siRNA. CDC2 siRNA was transfected as a positive control, as knockdown of CDC2 is known to affect the cell cycle and result in enlarged nuclei. Untransfected cells were also analyzed. After 72 hours, the nuclei of live cells were stained with Hoechst 33342 and the surface area of individual cell nuclei was measured using the cell^R Imaging System (Olympus). In this graph, 100 nuclear size measurements per treatment were plotted in order of size. AllStars Negative Control siRNA resulted in a nuclear size profile similar to untransfected cells. CDC2 siRNA resulted in enlarged nuclei as expected. The negative control siRNA, Control 1, also resulted in enlarged nuclei.Nuclear size phenotype unaffected.
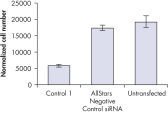
转染后细胞数量评估一系列负控制siRNAs是否正常细胞增殖。untransfected之间几乎没有区别在细胞数量观察细胞和细胞转染与消极的时候控制核。相比之下,转染后细胞数量显著减少与其他负控制siRNAs测试,如控制1,表明这些siRNAs导致增长缺陷表型(参见图”孔侑dF4y2Ba Identical numbers of MCF-7 cells were transfected with AllStars Negative Control siRNA or another negative control siRNA (Control 1). Untransfected cells were also analyzed. After 72 hours, cells were harvested, washed in PBS, transferred to TruCOUNT tubes, and counted using FACS analysis.Cell number unaffected.
核苷酸结合测量来确定DNA合成率untransfected hct - 116细胞,hct - 116细胞转染有一系列不同的负面siRNAs控制。核苷酸结合测量通过检查溴脱氧尿苷的吸收(BrdU),一个基地模拟胸苷的替代品胸苷在DNA复制,并纳入新合成的DNA。DNA合成速率的变化可能表明改变细胞生长和细胞周期。细胞转染与消极的时候控制核显示BrdU-incorporation率untransfected细胞非常相似。然而,另一个负控制核测试(1)控制导致了改变DNA合成水平较低,这表明该核影响细胞生长和细胞周期(参见图”孔侑dF4y2BaNormal DNA synthesis phenotype.
Live-cell染料排斥是用来测量细胞毒性的潜在影响的一系列负面siRNAs控制。结果表明,细胞转染与消极的时候控制核和untransfected细胞有类似的生活和死细胞的数量。相比之下,其他消极控制siRNAs导致细胞毒性的增加(见图”孔侑dF4y2BaNo increase in cytotoxic effects.
DNA染色后细胞固定是用来测量细胞循环分布(细胞的数量在G1 / G0,年代,和G2期细胞周期)。消极的时候控制核,转染后细胞的比例在每个阶段类似于观察untransfected细胞(参见图”孔侑dF4y2BaNormal cell-cycle distribution.
对准确负控制RNAi实验,消极控制核应该被纳入RISC (RNA-Induced沉默复杂)。这意味着控制核经过相同的生物过程gene-specific核,使比较的数据与数据从负控制siRNA gene-specific siRNA自信地判断结果,将目标基因击倒。孔侑dF4y2Ba
实验进行了如下。孔侑dF4y2Ba
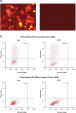
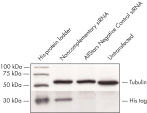
The reporter construct consisted of artificial target sequence complementary to AllStars Negative Control siRNA fused to a fluorescent reporter gene with a His tag.
Cotransfection noncomplementary siRNA导致的记者构造强烈表达荧光蛋白和他的标签。构造时cotransfected负控制siRNA时候,siRNA可拆卸的表达式的互补序列,导致整个退化mRNA转录本编码荧光报告基因,他的标签,可以阻止目标序列。融合蛋白的信使rna降解导致击倒(参见图”孔侑dF4y2Ba MCF-7 and HeLa cells were cotransfected with the reporter construct and either a noncomplementary siRNA or AllStars Negative Control siRNA. After 24 hours, expression of the fluorescent reporter gene was measured by [A] fluorescence microscopy (HeLa cells shown) and [B], [C] FACS analysis. Normal fluorescence was observed after cotransfection with the noncomplemetary siRNA, showing that the reporter gene is expressed. After cotransfection with AllStars Negative Control siRNA fluorescence was significantly decreased, showing that the reporter gene is downregulated. HeLa cells were cotransfected with the reporter construct and either a noncomplementary siRNA or AllStars Negative Control siRNA. After 48 hours, western blot analysis was used for measurement of expression of the His tag. The His tag was expressed after cotransfection with the noncomplementary siRNA. After cotransfection with AllStars Negative Control siRNA, the His tag was not detected.AllStars Negative Control siRNA is incorporated into RISC.
Western analysis shows AllStars Negative Control siRNA enters RISC.
HCT 116 cells were transfected with AllStars Negative Control siRNA, another negative control siRNA (Control 1), and CDC2 siRNA. CDC2 siRNA was transfected as a positive control, as knockdown of CDC2 is known to affect the cell cycle and result in enlarged nuclei. Untransfected cells were also analyzed. After 72 hours, the nuclei of live cells were stained with Hoechst 33342 and the surface area of individual cell nuclei was measured using the cell^R Imaging System (Olympus). In this graph, 100 nuclear size measurements per treatment were plotted in order of size. AllStars Negative Control siRNA resulted in a nuclear size profile similar to untransfected cells. CDC2 siRNA resulted in enlarged nuclei as expected. The negative control siRNA, Control 1, also resulted in enlarged nuclei.
Identical numbers of MCF-7 cells were transfected with AllStars Negative Control siRNA or another negative control siRNA (Control 1). Untransfected cells were also analyzed. After 72 hours, cells were harvested, washed in PBS, transferred to TruCOUNT tubes, and counted using FACS analysis.



The reporter construct consisted of artificial target sequence complementary to AllStars Negative Control siRNA fused to a fluorescent reporter gene with a His tag.

MCF-7 and HeLa cells were cotransfected with the reporter construct and either a noncomplementary siRNA or AllStars Negative Control siRNA. After 24 hours, expression of the fluorescent reporter gene was measured by [A] fluorescence microscopy (HeLa cells shown) and [B], [C] FACS analysis. Normal fluorescence was observed after cotransfection with the noncomplemetary siRNA, showing that the reporter gene is expressed. After cotransfection with AllStars Negative Control siRNA fluorescence was significantly decreased, showing that the reporter gene is downregulated.
HeLa cells were cotransfected with the reporter construct and either a noncomplementary siRNA or AllStars Negative Control siRNA. After 48 hours, western blot analysis was used for measurement of expression of the His tag. The His tag was expressed after cotransfection with the noncomplementary siRNA. After cotransfection with AllStars Negative Control siRNA, the His tag was not detected.
转染的负控制核每RNAi实验是必不可少的。结果从负控制应该与结果相比untransfected细胞。基因表达和表型理想情况下应该类似untransfected细胞和细胞转染与消极控制核。如果改变表达式或表型观察到细胞转染与消极控制核,这些变化是特异性的,他们是由于转染过程或核毒性而不是互补序列。非特异性效应应该最小,确保可靠RNAi / microrna的结果。孔侑dF4y2Ba
结果从负控制也可以与结果相比gene-specific核/ microrna的研究。这个比较允许研究者确定目标基因的影响可拆卸的基因表达和表型,因为消极的控制样本经历了相同的生物过程,惟一的差别是核/ microrna的序列。孔侑dF4y2Ba
结果从消极的时候控制核可以使用如下:孔侑dF4y2Ba
多个负控制siRNAs(控制1 - 10)在一式三份成MCF-7细胞转染。孵化后,cRNA准备和杂化Affymetrix人类U133 GeneChip数组。调节基因被确定为基因显示至少1.5倍变化表达式(调节和表达下调)相比untransfected细胞。创新途径分析软件被用来确定每个通路调控基因的比例相比,基因总数确定为中央通道。在酒吧出现在图中,这意味着由核基因通路的调控。如果每个通路基因调节,相对比例将为100%。低酒吧因此表明低调节基因通路内的相对比例。没有酒吧,没有由核基因通路的调控。时候负控制核(箭头)导致监管的最低数量的基因。相比之下,其他控制siRNAs导致更高的数字调节基因的重要的细胞通路。孔侑dF4y2Ba
| 特性孔侑dF4y2Ba | 规范孔侑dF4y2Ba |
|---|---|
| 设计孔侑dF4y2Ba | 预先设计/验证了Affymetrix GeneChip数组和基于单元的分析孔侑dF4y2Ba |
| 物种孔侑dF4y2Ba | 人类,老鼠,老鼠孔侑dF4y2Ba |
| 规模或收益孔侑dF4y2Ba | 5 nmol 20 nmol孔侑dF4y2Ba |
| 格式孔侑dF4y2Ba | 管孔侑dF4y2Ba |
| 目标序列提供了孔侑dF4y2Ba | 没有孔侑dF4y2Ba |
| 修改孔侑dF4y2Ba | 是的孔侑dF4y2Ba |